The model description so far is very much like a SEM model for multiple levels. In this chapter, we examine conventional random-slopes model for two level data. Mehta and West (2000) demonstrated how a longitudinal model with random-slopes for time may be estimated within an SEM context. Mehta, Neale, and Flay (2005) extended the idea to random-slopes model for the case of clustered data.
Table of Contents
Two-level equations
Level 1
\[ y_{ij} = b_{0j} + b_{1j} \times x_{ij} + e_{ij}, \] \[ e \sim N(0, \theta) \]
Level 2
\[ b_{0j} = \gamma_{00} + u_{0j} \] \[ b_{1j} = \gamma_{10} + u_{1j} \] \[ u \sim N(0, \Psi) \]
\Psi=
\begin{bmatrix}
\psi_{1,1} & \\
\psi_{2,1} & \psi_{2,2}
\end{bmatrix}
\]
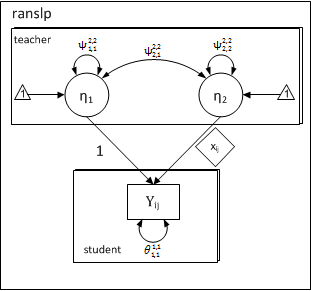
In a random-slopes model, the effect of a level-1 independent variable \( (x_{ij}) \) on a level-1 dependent variable \( (y_{ij}) \) varies randomly across level-2 units. Such models are also known as the random-coefficients model, as the regression-coefficients \( b_{0j} \) & \( b_{1j} \) vary randomly. Such random-effects are unobserved or latent variables at level-2. In xxM, random-effects are treated as latent variables. Using the SEM notation for a latent variable (\( \eta \)) the above equations may be written as:
\[ y_{ij}^1 = 1_{ij}^{1,2} \times \eta_{1j}^2 + x_{ij}^{1,2} \times \eta_{2j}^2 + e_{ij}^1, \]
\[ \eta_{1}^2 = \alpha_{1}^2 + \zeta_{1j}^2 \] \[ \eta_{2}^2 = \alpha_{2}^2 + \zeta_{2j}^2 \]
Note that the coefficients are random or latent variables, whereas the predictors \( (1_{ij} \) and \( x_{ij}) \) are fixed. We can use a SEM measurement model to capture the directional relationship between random-effects at level-2 and observed variables at level-1, in which the factor-loadings are fixed to level-1 values of fixed predictors. With this mind-set, the factor-loading matrix (\( \Lambda \)) has a single row and two columns:
\Lambda^{1,2} =
\begin{bmatrix}
1.0 & x_{ij}
\end{bmatrix}
\]
The first column is fixed to \( 1.0 \), whereas the second column is fixed to student-specific values of \( x_{ij} \). The idea of fixing factor-loadings for specifying multilevel random-slopes was introduced in the context of growth-curve modeling in Mehta and West (2000) and extended to clustered data and general multilevel case in Mehta, Neale, and Flay (2005). The following path-diagram has a one-to-one correspondence to the first and second level equations for a random-slope model.
Note:
- A random-slopes model is just a measurement model
Path Diagram

xxM Model Matrices
Level-1: Within matrices
In this example, we will use superscripts for associating each within-matrix to a given level. The rules of superscripts (for levels) correspond to the rules for subscripts (for variables).
Residual Covariance Matrix
At level-1, we have a single dependent variable and hence a single parameter level-1 residual variance (\( \theta_{1,1} \)). Hence, the residual covariance or theta matrix is a \( (1\times1) \) matrix:
\[ \Theta^{1,1} = [\theta_{1,1}^{1,1}] \]
The residual covariance matrix includes a superscript {1,1} to indicate that the matrix belongs to level-1. In principle it is possible to define covariance between variables across two different levels. For example, residual-covariance between the 3rd observed variable at level-5 and the 4th observed variable at level-2 may be represented as: \( \theta_{3,4}^{5,2} \). In this case, the superscripts indicate the levels of the respective variables in the subscript.
Level-2: Within Matrices
At level-2, we have two latent variables for the intercept and the slope and hence we have two latent means and a covariance matrix. In SEM, the latent variable mean vector is called alpha (\( \alpha \)) and the latent variable covariance matrix is called psi (\( \Psi \)).
Latent Means
With two latent variables, the latent variable mean matrix is a (2×1) matrix:
\( \alpha_{1}^{2} \) is the mean of the intercept and is the mean of the slope parameter or the average effect of \( x_{ij} \) on \( y_{ij} \). In the parlance of mixed-effects models, the latent means represent the fixed-effects of intercepts and slopes, respectively.
Latent Factor Covariance Matrix
Latent covariance matrix is a (2×2) matrix with two variances and a single covariance:
\Psi^{2,2}=
\begin{bmatrix}
\psi_{1,1}^{2,2} & \\
\psi_{2,1}^{2,2} & \psi_{2,2}^{2,2}
\end{bmatrix}
\]
\( \psi_{1,1}^{2,2} \) is the variance of the intercept factor representing variability in the intercept of \( y_{ij} \) across level-2 units and \( \psi_{2,2}^{2,2} \) is the variance of the slope parameter representing variability in the effect of \( x_{ij} \) on \( y_{ij} \). Finally, \( \psi_{2,1}^{2,2} \) is the covariance between the intercept and slope factors. In the parlance of mixed-effects models, the psi matrix represents the covariance among the random-effects.
Across level matrices: Teacher To Student
Across level factor-loading matrix
As described above, we need to capture the effect of level-2 intercept and slope factors on the level-1 dependent variable using a factor-loading matrix with fixed parameters. The factor-loading matrix \( (\Lambda^{1,2}) \) has a single row and two columns (1×2):
\[ \Lambda^{1,2} = \begin{bmatrix} \lambda_{1,1}^{1,2} & \lambda_{1,2}^{1,2} \end{bmatrix} \]
where,
\[ \lambda_{1,1}^{1,2}=1.0 \] and
\[ \lambda_{1,2}^{1,2}=x_{ij} \]
The first column is fixed to \( 1.0 \), whereas the second column is fixed to student-specific values of \( x_{ij} \). Note that we use superscripts to indicate that the factor-loading matrix connects latent variables at level-2 with observed variables at level-1. In fact, for an xxM model with multiple levels and observed and latent variables at each level, all model matrices require superscripts to uniquely associate the matrix with the level. In order to keep things simple, we have avoided superscripts for simple models considered so far. Very quickly it will become apparent that superscripts are necessary for our sanity.
Code Listing
xxM
The following code for the random-slopes model is nearly identical to the bivariate random-intercepts model presented earlier. In this case, there is a single level-1 dependent variable and a single level-1 independent variable. The number of matrices is the same as before; however, their dimensions are different as we have a single as opposed to two level-1 dependent variables. Practically, speaking the only new element is that the second factor-loading is fixed to \( x_{ij} \). This is accomplished by using an additional label matrix for the factor-loading matrix.
This is what we want to accomplish:
We do not wish to estimate any factor-loadings. Factor-loadings are fixed to \( 1.0 \) and \( x_{ij} \). Hence, both elements of pattern matrix are zero:
\[
\Lambda_{pattern} =
\begin{bmatrix}
0 & 0
\end{bmatrix}
\]
We want to fix the first factor-loading to 1.0 (intercept). We use the value matrix to provide the fixed-value of 1.0 for the first factor-loading. The second factor-loading does not have a single fixed value for every observation. Instead each observation (i) would have its own value for that factor-loading \( (x_{ij}) \). Clearly, the value matrix cannot be used for providing individual specific fixed values. Hence, the second element in the value matrix is left as 0.0. xxM ignores it internally.
\[
\Lambda_{value} =
\begin{bmatrix}
1.0 & 0.0
\end{bmatrix}
\]
The job of fixing the factor-loading is left to the label matrix. A label matrix is used to assign labels to each parameter within the matrix. Label matrices can be used to impose equality constraints across matrices. Any two parameters with the same label are constrained to be equal. Label matrices are also used for specifying that a specific parameter is to be fixed to data-values. In this case, the first label is irrelevant as that parameter has already been fixed to 1.0. We use a descriptive label “lambda11” as the first label (something such as Justin would have worked as well). The second factor-loading is the one we are interested in. We want to fix the second factor-loading to the observation specific value of the predictor \( (x_{ij}) \). This is accomplished by using a two-part label: levelName.predictorName. In this case, the predictor is a student level variable. Hence, the first part of the label is student. The second part is the actual predictor name, in this case x.
\[
\Lambda_{label} =
\begin{bmatrix}
“dog” & “student.x”
\end{bmatrix}
\]
lambda_label <- matrix(c("lambda11", "student.x"), 1, 2)
The complete listing of xxM code for the random-slopes example follows:
Load xxM
xxM library needs to be loaded first. Data for the model must be available in the workspace.
library(xxm)
data(pcwa.xxm, package = "xxm")
Construct R-matrices
For each parameter matrix, construct three related matrices:
- pattern matrix: A matrix indicating free or fixed parameters.
- value matrix: with start or fixed values for corresponding parameters.
- label matrix: with user friendly label for each parameter. label matrix is optional.
alpha_pattern <- matrix(1, 2, 1)
alpha_value <- (matrix(c(443, 1), 2, 1))
psi_pattern <- matrix(1, 2, 2)
psi_value <- (matrix(c(10, 0, 0, 0.05), 2, 2))
lambda_pattern <- matrix(c(0, 0), 1, 2)
lambda_value <- (matrix(c(1, 0), 1, 2))
lambda_label <- (matrix(c("l11", "student.wa"), 1, 2))
theta_pattern <- matrix(1, 1, 1)
theta_value <- (matrix(200, 1, 1))
Construct model
xxmModel() is used to declare level names. The function returns a model object that is passed as a parameter to subsequent statements. Variable name for the return value can be abything. A
ranslp <- xxmModel(levels = c("student", "teacher"))
## A new model was created.
Add submodels
For each declared level xxmSubmodel() is invoked to add corresponding submodel to the model object. The function adds three types of information to the model object:
- parents declares a list of all parents of the current level.
- Level with the independent variable is the parent level.
- Level with the dependent variable is the child level.
- variables declares names of observed dependent (ys), observed independent (xs) and latent variables (etas) for the level.
- data R data object for the current level.
### Submodel: Student
ranslp <- xxmSubmodel(model = ranslp, level = "student", parents = "teacher",
ys = "pc", xs = c("wa"), etas = , data = pcwa.student)
## Submodel for level `student` was created.
### Submodel: School
ranslp <- xxmSubmodel(model = ranslp, level = "teacher", parents = , ys = ,
xs = , etas = c("int", "slp"), data = pcwa.teacher)
## Submodel for level `teacher` was created.
Add within-level matrices
For each declared level xxmWithinMatrix() is used to add within-level parameter matrices. For each parameter matrix, the function adds the three matrices constructed earlier:
- pattern
- value
- label (optional)
# Level 1: Student model
ranslp <- xxmWithinMatrix(model = ranslp, level = "student", type = "theta",
pattern = theta_pattern, value = theta_value, , )
##
## 'theta' matrix does not exist and will be added.
## Added `theta` matrix.
# Level 2: Teacher model
ranslp <- xxmWithinMatrix(model = ranslp, level = "teacher", type = "alpha",
pattern = alpha_pattern, value = alpha_value, , )
##
## 'alpha' matrix does not exist and will be added.
## Added `alpha` matrix.
ranslp <- xxmWithinMatrix(model = ranslp, level = "teacher", type = "psi", pattern = psi_pattern,
value = psi_value, , )
##
## 'psi' matrix does not exist and will be added.
## Added `psi` matrix.
Add across-level matrices
Pairs of levels that share parent-child relationship have regression relationships. xxmBetweenMatrix() is used to add corresponding rergession matrices connecting the two levels.
- Level with the independent variable is the parent level.
- Level with the dependent variable is the child level.
For each parameter matrix, the function adds the three matrices constructed earlier:
- pattern
- value
- label (optional)
# Teacher -> Student model
ranslp <- xxmBetweenMatrix(model = ranslp, parent = "teacher", child = "student",
type = "lambda", pattern = lambda_pattern, value = lambda_value, label = lambda_label,
)
##
## 'lambda' matrix does not exist and will be added.
## Added `lambda` matrix.
Estimate model parameters
Estimation process is initiated by xxmRun(). If all goes well, a q&d summary of the results is printed.
ranslp <- xxmRun(ranslp)
## ------------------------------------------------------------------------------
## Estimating model parameters
## ------------------------------------------------------------------------------
## 6784.1745837625
## 6778.1391075981
## 6773.1200961903
## 6762.9332973861
## 6760.2695123117
## 6755.3346517174
## 6754.7166699919
## 6754.6488166234
## 6754.6345238311
## 6754.6252303872
## 6754.6251990707
## 6754.6251984028
## 6754.6251983926
## Model converged normally
## nParms: 6
## ------------------------------------------------------------------------------
## *
## 1: student_theta_1_1 :: 234.559 [ 0.000, 0.000]
##
## 2: teacher_psi_1_1 :: 37.099 [ 0.000, 0.000]
##
## 3: teacher_psi_1_2 :: -0.340 [ 0.000, 0.000]
##
## 4: teacher_psi_2_2 :: 0.042 [ 0.000, 0.000]
##
## 5: teacher_alpha_1_1 :: 443.386 [ 0.000, 0.000]
##
## 6: teacher_alpha_2_1 :: 0.850 [ 0.000, 0.000]
##
## ------------------------------------------------------------------------------
Estimate profile-likelihood confidence intervals
Once parameters are estimated, confidence intervals are estimated by invoking xxmCI(). Depending on the the number of observations and the complexity of the model, xxmCI() may take a long time to compute. xxmCI() also prints a summary of parameter estimates and CIS.
ranslp <- xxmCI(ranslp)
View results
A summary of results may be retrived as an R list by a call to xxmSummary(). The returned list has two elements:
- fit is a list with five elements:
- deviance is \( -2 Log Likelihood \) for the maximum likelihood fit function.
- nParameters is the total number of unique parameters.
- nObservations is the total number of observations across all levels.
- aic is Akaike’s Information Criterion or AIC computed as \( -2ll + 2*p \).
- bic is Bayesian Information Criterion or BIC computed as \( -2ll + p*\log(n) \).
- estimates is a single table of free parameter estimates
All xxM parameters have superscripts {child, parent} and subscripts {to, from}. xxM adds a descriptive parameter label if one is not already provided by the user.
xxmSummary(ranslp)
## $fit
## $fit$deviance
## [1] 6755
##
## $fit$nParameters
## [1] 6
##
## $fit$nObservations
## [1] 802
##
## $fit$aic
## [1] 6767
##
## $fit$bic
## [1] 6795
##
##
## $estimates
## child parent to from label estimate low high
## 1 student student pc pc student_theta_1_1 234.55860 211.014004 261.5897
## 4 teacher teacher int int teacher_psi_1_1 37.09872 19.563135 64.1247
## 5 teacher teacher int slp teacher_psi_1_2 -0.34001 -1.111982 0.3339
## 6 teacher teacher slp slp teacher_psi_2_2 0.04239 0.006877 0.1010
## 7 teacher teacher int One teacher_alpha_1_1 443.38580 441.593620 445.1855
## 8 teacher teacher slp One teacher_alpha_2_1 0.85014 0.765115 0.9395
Free model object
xxM model object may hog a significant amount of RAM outside of R’s workspace. This memory will automatically be released, when the workspace is cleared by a call to rm(list=ls()) or at the end of the R session. Alternatively, it is recommended that xxmFree() may be called to release the memory.
ranslp <- xxmFree(ranslp)
The above code appears to be very complicated. However, as the model becomes increasingly complex with multiple levels, variables, and constraints, the xxM code will remain succinct. Thus, although there is an overhead for simple models, for complex models the matrix notation is a blessing.
Proc Mixed
Proc Mixed data = ranslp covtest;
CLASS teacher;
MODEL y = x/s;
RANDOM Intercept x/subject = teacher G type = UN;
RUN;
SAS code for a random-slopes model uses a CLASS statement to identify the level-2 units, in this case teacher. The MODEL statement estimates the fixed-effects (\( \alpha \)). The RANDOM statement specifies that the level-1 intercepts and the effect of level-1 predictor \( x_{ij} \) is allowed to vary across “subject” (i.e., the level-2 units). The covariance among the random-effects is freely estimated (specified by a “type = UN”) and the covariance matrix is referred to as the G matrix. The \( G \) matrix corresponds to the xxM \( \Psi \) matrix. Finally, like all regression models, Proc Mixed estimates the residual variance of level-1 dependent variable (\( \theta_{1,1} \)) by default. The important thing to note is that there is one-to-one correspondence between the parameters estimated in Proc Mixed and SEM.
R: LME4
lmer(y ~ 1 + x + (1 + x | teacher), ranslp)
For the current dataset, the parameter estimates are: