Table of Contents
TWO LEVEL CONFIRMATORY FACTOR ANALYSIS
The present example combines aspects of the models in chapter 3 (random slopes model) and chapter 7 (two-level confirmatory factor analysis).
Motivating Example
Data were simulated for a population model with five observed variables at level-1, four of which serve as indicators for latent factors at levels 1 and 2. The level-1 latent construct is regressed on the remaining observed variable and this coefficient is allowed to vary randomly across level-2 units (i.e. random slope).
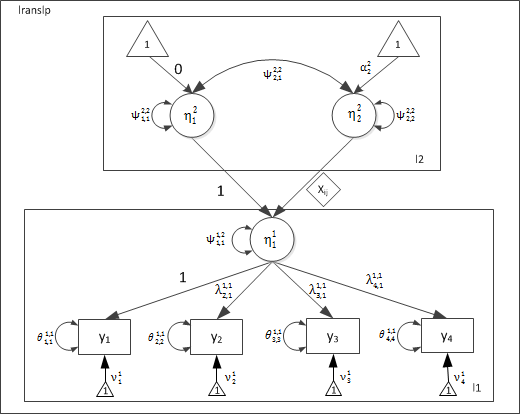
Conditional Two-Level CFA Model with a random slope
As in the previous example, each observed indicator (y1-y4) varies across levels 1 and 2, and these indicators are correlated within each level. Estimating a common latent factor at each level of analysis allows us to parsimoniously explain the common variance/covariance among the indicators. Moreover, the latent factor at level-1 \( (\eta_1^1) \) is regressed on the exogenous observed predictor (x1). This effect varies randomly at level-2, leading to two latent variables at level-2, corresponding to the correlated intercept \( (\eta_1^2) \) and slope \( (\eta_2^2) \) factors.
Path Diagram
Scalar Representation
Level-1 model
The measurement model for each level-1 indicator can be presented as:
\[ y_{pij}^1= \nu_p^1 + \lambda_p^{1,1} \times \eta_{1ij}^1 + e_{pij}^1 \]
where, \( y_{pij} \) is \( p^{th} \) observed indicator for observation \( i \), nested within cluster \( j \).
\[ \eta_{1ij}^1 = \beta_{1,1}^{1,2} \times \eta_{1j}^2 + \beta_{1,2}^{1,2}\times \eta_{2j}^2 + \xi_{1ij}^1 \]
\[ \xi_{1ij}^1 \sim N(0,\psi_{1,1}^{1,1}) \] \[ e_{pij}^1 \sim N(0,\theta^{1,1}) \]
The level 1 model hypothesizes following parameters:
- \( (p-1) \) factor loadings \( (\lambda_p) \) . The first factor-loading is fixed to 1.0 for scale identification.
- Residual variance for each of the p observed indicators \( (\theta_{p,p}) \).
- Latent-on-Latent regression coefficients (\( \beta_{1,1}^{1,2} \) & \( \beta_{1,2}^{1,2} \)) linking the level-2 random intercept \( (\eta_{1j}^2) \) and slope \( (\eta_{2j}^2) \) to the level-1 latent factor \( (\eta_{1ij}^1) \).
- A single latent residual variance \( (\psi_{1,1}^{1,1}) \). Note that \( \eta_{1ij}^1 \) is influenced by \( \eta_{1j}^2 \) and \( \eta_{2j}^2 \) at level 2.
- Measurement intercepts for each of the p observed indicators \( (\nu_p) \).
Level-2 Levels
The teacher level has a two latent variables corresponding to the random intercept \( (\eta_1^2) \) and slope \( (\eta_1^2) \):
\[ \eta_1^2 \sim N(\alpha_1^2,\psi_{1,1}^{2,2} ) \]
\[ \eta_2^2 \sim N(\alpha_2^2,\psi_{2,2}^{2,2} ) \]
XXM Model Matrices
Level-1
Factor-loading matrix (Lambda)
\Lambda_{pattern}^{1,1} =
\begin{bmatrix}
0 \\
1 \\
1 \\
1
\end{bmatrix}
\]
\Lambda_{value}^{1,1} =
\begin{bmatrix}
1.0 \\
1.1 \\
0.9 \\
0.8
\end{bmatrix}
\]
First factor loading is fixed to 1.0. Hence, we need to fix the first parameter in the pattern matrix. The value at which it is being fixed to is specified in the value matrix. In this case the first factor-loading is being fixed to a value of 1.0.
Observed Residual Covariance Matrix (Theta)
\Theta_{pattern}^{1,1} =
\begin{bmatrix}
1 & 0 & 0 & 0 \\
0 & 1 & 0 & 0 \\
0 & 0 & 1 & 0 \\
0 & 0 & 0 & 1
\end{bmatrix}
\]
\Theta_{value}^{1,1} =
\begin{bmatrix}
1.1 & 0.0 & 0.0 & 0.0 \\
0.0 & 2.1 & 0.0 & 0.0 \\
0.0 & 0.0 & 1.3 & 0.0 \\
0.0 & 0.0 & 0.0 & 1.5
\end{bmatrix}
\]
Residual covariance matrix is a diagonal matrix, meaning we are only estimating residual variances. Residual covariances are all fixed to 0. Again we use a pattern and a value matrix to “fix” all off-diagonal elements to 0.0.
Latent Residual Covariance Matrix (PSI)
\Psi_{pattern}^{1,1} =
\begin{bmatrix}
1
\end{bmatrix}
\]
\Psi_{value}^{1,1} =
\begin{bmatrix}
1.1
\end{bmatrix}
\]
Observed Variablel Intercepts (nu)
\nu_{pattern}^{1}=
\begin{bmatrix}
1 \\
1 \\
1 \\
1
\end{bmatrix}
\]
,
\nu_{value}^{1}=
\begin{bmatrix}
1.1 \\
2.1 \\
1.3 \\
0.7
\end{bmatrix}
\]
Latent on Latent regression coefficient matrix (Beta)
B_{pattern}^{1,2}=
\begin{bmatrix}
0 & 1
\end{bmatrix}
\]
,
B_{value}^{1,2}=
\begin{bmatrix}
1.0 & 0.0 \\
\end{bmatrix}
\]
The level-1 latent variable is regressed on the intercept and slope factors at level-2. Estimating a random slope for the level-1 predictor X requires that we specify a label matrix \( B_{label}^{1,2} \) that tells xxM to use subject-specific values for the regression of \( \eta_1^1 \) on X.
B_{label}^{1,2}=
\begin{bmatrix}
“dog” & “l1.X” \\
\end{bmatrix}
\]
The first element of the label matrix (1, 1) corresponds to the regression of \( \eta_1^1 \) on \( \eta_1^2 \), which identifies the level-1 random intercept for the latent factor, and the second element (1, 2) corresponds to the regression of \( \eta_1^1 \) on \( \eta_2^2 \), which identifies the random slope of \( \eta_1^1 \) on X. The label provided in the first element is arbitrary, and could have just as well been fido, cat, or any other character string. In contrast, the label provided for the second element (corresponding to the random slope) is not arbitrary, as it must indicate the name of the lower-level predictor dataset and variable in the format: datasetName.variableName. The presence of a period ( . ) in the label matrix causes xxM to search the specified dataset for the predictor variable and insert observation-specific values for the factor-loading. This specification identifies the random slope factor \( \eta_2^2 \), which allows the regression of \( \eta_1^1 \) on X to vary across level-2 subjects. In the current example, the value \( \beta_{1,2}^{1,2} = “l1.X” \) tells xxM to estimate a random slope for variable X found in dataset \( l1 \).
Level-2
Latent Covariance Matrix (Psi)
\Psi_{pattern}^{2,2}=
\begin{bmatrix}
0 \\
1 & 1
\end{bmatrix}
\]
\Psi_{value}^{2,2}=
\begin{bmatrix}
0.1 \\
0.0 & 0.1
\end{bmatrix}
\]
,
There are only latent variables at level-2, corresponding to the intercept and slope factors.
Latent Mean Matrix
\alpha_{pattern}^2=
\begin{bmatrix}
0 \\
1
\end{bmatrix}
\]
\alpha_{value}^2=
\begin{bmatrix}
0.0 \\
0.5
\end{bmatrix}
\]
,
Mean structure for the random intercept is modeled at level-1, therefore the level-2 intercept for level-1 latent variable \( (\alpha_1^2) \) is not identified and must be fixed to 0.0. \( \alpha_2^2 \) represents the mean of the slope factor, or the fixed regression coefficient for \( X \).
Model Matrices Summary
The following table provides a complete summary of parameter matrices:
| Type | Matrix | Pattern |
|---|---|---|
| level 1: \( \Theta \) |
\[
\Theta^{1,1} = \begin{bmatrix} \theta_{1,1}^{1,1} & \\ \theta_{2,1}^{1,1} & \theta_{2,2}^{1,1} \\ \theta_{3,1}^{1,1} & \theta_{3,2}^{1,1} & \theta_{3,3}^{1,1} \\ \theta_{4,1}^{1,1} & \theta_{4,2}^{1,1} & \theta_{4,3}^{1,1} & \theta_{4,4}^{1,1} \\ \end{bmatrix} \] |
\[
\Theta^{1,1} = \begin{bmatrix} \ 1 \\ \ 0 & 1 \\ \ 0 & 0 & 1 \\ \ 0 & 0 & 0 & 1\\ \end{bmatrix} \] |
| level 1: \( \nu \) |
\[
\nu^{1} = \begin{bmatrix} \nu_1^1\\ \nu_2^1\\ \nu_3^1 \\ \nu_4^1 \end{bmatrix} \] |
\[
\nu^{1} = \begin{bmatrix} 1\\ 1\\ 1\\ 1 \end{bmatrix} \] |
| level 1: \( \Lambda \) |
\[
\Lambda^{1,1} = \begin{bmatrix} \lambda_{1,1}^{1,1}\\ \lambda_{2,1}^{1,1}\\ \lambda_{3,1}^{1,1}\\ \lambda_{4,1}^{1,1} \end{bmatrix} \] |
\[
\Lambda^{1,1} = \begin{bmatrix} 0\\ 1\\ 1\\ 1 \end{bmatrix} \] |
| level1: \( \Psi \) |
\[
\Psi^{1,1} = [\psi_{1,1}^{1,1}] \] |
\[
\Psi^{1,1} = [1] \] |
| Level 2 -> Level 1: \( B \) |
\[
B^{1,2} = \begin{bmatrix} \beta_{1,1}^{1,2} & \beta_{1,2}^{1,2}\\ \end{bmatrix} \] |
\[
B^{1,2} = \begin{bmatrix} 0 & 1\\ \end{bmatrix} \] |
| level 2: \( \Psi \) |
\[
\Psi^{1,2} = \begin{bmatrix} \psi_{1,1}^{2,2} &\\ \psi_{2,1}^{2,2} & \psi^{2,2}_{2,2}\\ \end{bmatrix} \] |
\[
\Psi^{1,2} = \begin{bmatrix} 1 & \\ 1 & 1 \\ \end{bmatrix} \] |
| level 2: \( \alpha \) |
\[
\alpha^{1,2} = \begin{bmatrix} \alpha_1^2 \\ \alpha_2^2 \end{bmatrix} \] |
\[
\alpha^{1,2} = \begin{bmatrix} 0 \\ 1 \end{bmatrix} \] |
Code Listing
xxM
Load xxM and data
library(xxm)
data(lranslp.xxm)
Construct R-matrices
For each parameter matrix, construct three related matrices:
- pattern matrix: A matrix indicating free or fixed parameters.
- value matrix: with start or fixed values for corresponding parameters.
- label matrix: with user friendly label for each parameter. label matrix is optional.
# l1: Factor-Loading Matrix
ly1_pat <- matrix(c(0, 1, 1, 1), 4, 1)
ly1_val <- matrix(c(1, 1, 1, 1), 4, 1)
# l1:latent variable-covariance matrix
ps1_pat <- matrix(1, 1, 1)
ps1_val <- matrix(1, 1, 1)
# l1:observed residual variable-covariance matrix
th1_pat <- matrix(c(1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 1), 4, 4, byrow = TRUE)
th1_val <- matrix(c(1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 1), 4, 4, byrow = TRUE)
# l1: observed variable intercepts
nu1_pat <- matrix(c(1, 1, 1, 1), 4, 1)
nu1_val <- matrix(c(0.9, 0.7, 0.7, 0.6), 4, 1)
# l2: latent variable covariance matrix
ps2_pat <- matrix(1, 2, 2)
ps2_val <- matrix(c(0.2, 0.01, 0.01, 0.2), 2, 2)
# l2: intercepts for level 2 slope variables
al2_pat <- matrix(c(0, 1), 2, 1)
al2_val <- matrix(c(0, 0.2), 2, 1)
# l2 -> l1 factor loading matrix
be12_pat <- matrix(c(0, 0), 1, 2)
be12_val <- matrix(c(1, 0), 1, 2)
be12_label <- matrix(c("one", "l1.x"), 1, 2)
Construct main model object
xxmModel() is used to declare level names. The function returns a model object that is passed as a parameter to subsequent stattements.
lranslp <- xxmModel(levels = c("l1", "l2"))
## A new model was created.
Add submodels to the model objects
For each declared level xxmSubmodel() is invoked to add corresponding submodel to the model object. The function adds three pieces of information:
1. parents declares a list of parents of the current level.
2. variables declares names of observed dependent (ys), observed independent (xs) and latent variables (etas) for the level.
3. data R data object for the current level.
### Submodel: l1
lranslp <- xxmSubmodel(model = lranslp, level = "l1", parents = "l2", ys = c("y1",
"y2", "y3", "y4"), xs = "x", etas = "fw", data = l1)
## Submodel for level `l1` was created.
### Submodel: l2
lranslp <- xxmSubmodel(model = lranslp, level = "l2", parents = , ys = , xs = ,
etas = c("int", "slp"), data = l2)
## Submodel for level `l2` was created.
Add Within-level parameter matrices for each submodel
For each declared level xxmWithinMatrix() is used to add within-level parameter matrices. For each parameter matrix, the function adds the three matrices constructed earlier:
- pattern
- value
- label (optional)
# l1 within matrices (lambda, psi, theta)
lranslp <- xxmWithinMatrix(model = lranslp, level = "l1", type = "lambda", pattern = ly1_pat,
value = ly1_val)
##
## 'lambda' matrix does not exist and will be added.
## Added `lambda` matrix.
lranslp <- xxmWithinMatrix(model = lranslp, level = "l1", type = "psi", pattern = ps1_pat,
value = ps1_val)
##
## 'psi' matrix does not exist and will be added.
## Added `psi` matrix.
lranslp <- xxmWithinMatrix(model = lranslp, level = "l1", type = "theta", pattern = th1_pat,
value = th1_val)
##
## 'theta' matrix does not exist and will be added.
## Added `theta` matrix.
lranslp <- xxmWithinMatrix(model = lranslp, level = "l1", type = "nu", pattern = nu1_pat,
val = nu1_val)
##
## 'nu' matrix does not exist and will be added.
## Added `nu` matrix.
# l2 within matrices (psi, alpha)
lranslp <- xxmWithinMatrix(model = lranslp, level = "l2", type = "psi", pattern = ps2_pat,
value = ps2_val)
##
## 'psi' matrix does not exist and will be added.
## Added `psi` matrix.
lranslp <- xxmWithinMatrix(model = lranslp, level = "l2", type = "alpha", pattern = al2_pat,
value = al2_val)
##
## 'alpha' matrix does not exist and will be added.
## Added `alpha` matrix.
Add Across-level parameter matrices to the model
Pairs of levels that share parent-child relationship have regression relationships. xxmBetweenMatrix() is used to add corresponding parameter matrices connecting the two levels.
- Level with the independent variable is the parent level.
- Level with the dependent variable is the child level.
For each parameter matrix, the function adds the three matrices constructed earlier:
- pattern
- value
- label (optional)
## l2->l1 loading matrix (beta)
lranslp <- xxmBetweenMatrix(model = lranslp, parent = "l2", child = "l1", type = "beta",
pattern = be12_pat, value = be12_val, label = be12_label)
##
## 'beta' matrix does not exist and will be added.
## Added `beta` matrix.
Estimate model parameters
Estimation process is initiated by xxmRun(). If all goes well, a quick printed summary of results is produced.
lranslp <- xxmRun(lranslp)
## ------------------------------------------------------------------------------
## Estimating model parameters
## ------------------------------------------------------------------------------
## 2503.7965065730
## 2498.3032714776
## 2497.5686604366
## 2496.4932421838
## 2495.2162015696
## 2493.7499666352
## 2491.9162316733
## 2489.5589752111
## 2488.5926066982
## 2486.4234202193
## 2484.8218001901
## 2484.1568716505
## 2482.5926264028
## 2479.4619807727
## 2478.3347722737
## 2475.5559374325
## 2475.0332077759
## 2474.1785810527
## 2474.0723191035
## 2473.7506485369
## 2473.6260469653
## 2473.6220281160
## 2473.6210235790
## 2473.6209732651
## 2473.6209668752
## 2473.6209667384
## 2473.6209667317
## Model converged normally
## nParms: 16
## ------------------------------------------------------------------------------
## *
## 1: l1_theta_1_1 :: 1.205 [ 0.000, 0.000]
##
## 2: l1_theta_2_2 :: 0.929 [ 0.000, 0.000]
##
## 3: l1_theta_3_3 :: 1.227 [ 0.000, 0.000]
##
## 4: l1_theta_4_4 :: 1.402 [ 0.000, 0.000]
##
## 5: l1_psi_1_1 :: 0.706 [ 0.000, 0.000]
##
## 6: l1_nu_1_1 :: 0.914 [ 0.000, 0.000]
##
## 7: l1_nu_2_1 :: 0.665 [ 0.000, 0.000]
##
## 8: l1_nu_3_1 :: 0.623 [ 0.000, 0.000]
##
## 9: l1_nu_4_1 :: 0.453 [ 0.000, 0.000]
##
## 10: l1_lambda_2_1 :: 1.048 [ 0.000, 0.000]
##
## 11: l1_lambda_3_1 :: 0.794 [ 0.000, 0.000]
##
## 12: l1_lambda_4_1 :: 0.735 [ 0.000, 0.000]
##
## 13: l2_psi_1_1 :: 0.600 [ 0.000, 0.000]
##
## 14: l2_psi_1_2 :: -0.214 [ 0.000, 0.000]
##
## 15: l2_psi_2_2 :: 0.308 [ 0.000, 0.000]
##
## 16: l2_alpha_2_1 :: 0.255 [ 0.000, 0.000]
##
## ------------------------------------------------------------------------------
Estimate profile-likelihood confidence intervals
Once parameters are estimated, confidence inetrvals are estimated by invoking xxmCI() . Depending on the the number of observations and the complexity of the dependence structure xxmCI() may take very long. xxMCI() displays a table of parameter estimates and CIS.
View results
A summary of results may be retrived as an R list by a call to xxmSummary()
summary <- xxmSummary(lranslp)
summary
## $fit
## $fit$deviance
## [1] 2474
##
## $fit$nParameters
## [1] 16
##
## $fit$nObservations
## [1] 716
##
## $fit$aic
## [1] 2506
##
## $fit$bic
## [1] 2579
##
##
## $estimates
## child parent to from label estimate low high
## 1 l1 l1 y1 y1 l1_theta_1_1 1.2047 0.83409 1.66413
## 3 l1 l1 y2 y2 l1_theta_2_2 0.9287 0.58123 1.33529
## 6 l1 l1 y3 y3 l1_theta_3_3 1.2266 0.92552 1.60449
## 10 l1 l1 y4 y4 l1_theta_4_4 1.4022 1.07819 1.81434
## 11 l1 l1 fw fw l1_psi_1_1 0.7065 0.40596 1.14151
## 12 l1 l1 y1 One l1_nu_1_1 0.9136 0.60178 1.22260
## 13 l1 l1 y2 One l1_nu_2_1 0.6654 0.36756 0.96239
## 14 l1 l1 y3 One l1_nu_3_1 0.6226 0.36733 0.87836
## 15 l1 l1 y4 One l1_nu_4_1 0.4533 0.20020 0.70703
## 17 l1 l1 y2 fw l1_lambda_2_1 1.0484 0.82693 1.33778
## 18 l1 l1 y3 fw l1_lambda_3_1 0.7935 0.61057 1.02356
## 19 l1 l1 y4 fw l1_lambda_4_1 0.7353 0.54762 0.96547
## 22 l2 l2 int int l2_psi_1_1 0.6003 0.30858 1.12230
## 23 l2 l2 int slp l2_psi_1_2 -0.2141 -0.50183 -0.02663
## 24 l2 l2 slp slp l2_psi_2_2 0.3078 0.12205 0.65693
## 26 l2 l2 slp One l2_alpha_2_1 0.2555 0.03513 0.48442
Free model object
xxM model object may hog a large amount of RAM outside of R’s memory. This memory will automatically be released, when R’s workspace is cleared by a call to rm(list=ls()) or at the end of the R session. Alternatively, xxmFree() may be called to release memory.
lranslp <- xxmFree(lranslp)
For the current dataset, the parameter estimates are:
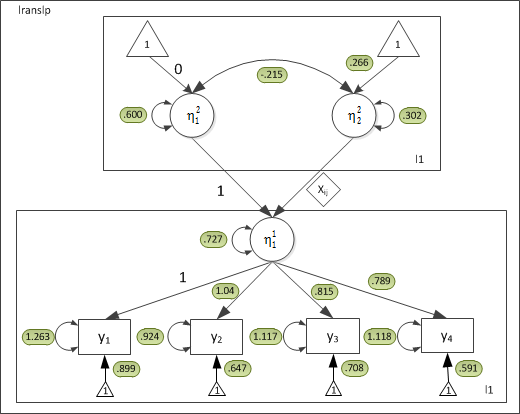
Because constrained the residual variances to be equal, all the thetas will be the same.