This tutorial introduces core concepts of xxM as a modeling framework and as a software package. Key elements of model specification in xxM are introduced in the context of fitting a bivariate random-intercepts model (Mehta, Neale, & Flay, 2005). Although the example is relatively trivial, once you understand the building blocks presented in this tutorial, you should be able to construct complex models easily.
The presentation is in three sections:
- Bivariate random-intercepts model: Representation using four different perspectives.
- Multilevel modeling (MLM)
- xxM
- Linear mixed-effects (LME) model
- Path diagram
- Description of the process and steps of fitting the model in xxM.
- Code listing
- xxM: An annoated summary of a session of fitting the model
- SAS Proc Mixed
Table of Contents
Bivariate random-intercepts model
MLM
Level 1
\[ y_{pij} = 1 \times \eta_{pj} + e_{pij}, e \sim N(0, \Theta),\]
where subscripts \( p \), \( i \) and \( j \) correspond to the variable, subject and cluster, respectively. \( y_{pij} \) is the \( p^{th} \) dependent variable and \( \eta_{pj} \) is the corresponding random-intercept. Residuals are assumed to be distributed normally with covariance,
\Theta =
\begin{bmatrix}
\theta_{1,1} & \\
\theta_{2,1} & \theta_{2,2}
\end{bmatrix}.
\]
Level 2
Random intercepts or level-2 latent variables are distributed normally,
\[ \eta_{pj} = \alpha_{p} + u_{pj}, u \sim N(0, \Psi), \]
\Psi=
\begin{bmatrix}
\psi_{1,1} & \\
\psi_{2,1} & \psi_{2,2}
\end{bmatrix},
\alpha=
\begin{bmatrix}
\alpha_{1} \\
\alpha_{2}
\end{bmatrix}.
\]
The bivariate random-intercepts model has 8 parameters:
- Covariance among level-1 residuals \( (e_{pij}) \), denoted as \( \theta_{21} \) in \( \Theta \) matrix.
- Covariance among level-2 random-intercepts \( (u_{pj}) \) , denoted as \( \psi_{21} \) in \( \Psi \) matrix.
- Grand-means of \( y_1 \) and \( y_2 \), denoted as \( \alpha_1 \) and \( \alpha_2 \) in \( \alpha \) vector.
- Variances of level-1 residuals, denoted as \( \theta_{11} \) and \( \theta_{22} \) in \( \Theta \) matrix.
- Variances of the level-2 random-intercepts, denoted as \( \psi_{11} \) and \( \psi_{22} \) in \( \Psi \) matrix.
- Factor-loading matrix of \( \Lambda \) is an identity matrix and does not include any free parameters.
xxM
xxM uses a different approach to represent NL-SEM model.
\[ y_i^{1} = \Lambda_{i,j}^{1,2} \times \eta_j^{2} + e_{i,j}^{1}. \] The equation applies to generic units \( i \) and \( j \) at each level. Hence, subscripts are not really necessary. These are included to make connection with the MLM representation obvious.
Observed and latent observations
For a generic unit, the multivariate vector of observations are:
\begin{bmatrix}
y_{1}^{1} \\
y_{2}^{1}
\end{bmatrix},
e^{1}=
\begin{bmatrix}
e_{1}^{1} \\
e_{2}^{1}
\end{bmatrix}
\text{, & } \eta^{2}=
\begin{bmatrix}
\eta_{1}^{2} \\
\eta_{2}^{2}
\end{bmatrix}.
\]
There are three advantages to using a superscript indicating levels:
- Subscripts are no longer crowded with separate identifiers for each level.
- In fact, as the equations always apply to generic units, subject related subscripts are generally unnecessary!
- Model specification can be done using compact matrices.
xxM model matrices
With vector of observations defined as above, the meaning of superscripts and subscripts in corresponding model marices is self-evident.
\Theta^{1,1} =
\begin{bmatrix}
\theta_{1,1}^{1,1} & \\
\theta_{2,1}^{1,1} & \theta_{2,2}^{1,1}
\end{bmatrix}
,
\Psi^{2,2}=
\begin{bmatrix}
\psi_{1,1}^{2,2} & \\
\psi_{2,1}^{2,2} & \psi_{2,2}^{2,2}
\end{bmatrix}
,
\alpha^{2}=
\begin{bmatrix}
\alpha_{1}^{2} \\
\alpha_{2}^{2}
\end{bmatrix}.
\]
For two dependent variables, we can write MLM equation as:
\[ y_{1ij} = 1 \times \eta_{1j} + 0 \times \eta_{2j} + e_{1ij} \text{, &} \] \[ y_{2ij} = 0 \times \eta_{1j} + 1 \times \eta_{2j} + e_{2ij}. \]
Notice that the regression coefficients of the random-effects are fixed to 0.0 or 1.0. We gather these coefficients into a single matrix \( \Lambda \)
\Lambda =
\begin{bmatrix}
\lambda_{1,1}^{1,2} & \lambda_{1,1}^{1,2} \\
\lambda_{2,1}^{1,2} & \lambda_{2,2}^{1,2}
\end{bmatrix}
=
\begin{bmatrix}
1.0 & 0.0\\
0.0 & 1.0
\end{bmatrix}.
\]
Why xxM representation?
MLM specification of any and all models involve superscripts referring to variables, levels, and generic units at each level. This leads to subscript explosion even for simple models. In contrast, xxM orthogonalizes information about variables, levels and units within levels. As a result, xxM specification remains the same – regardless of the number of levels or the complexity of the dependency structure (hierarchical or cross-classified or mixed)!
xxM model matrix specification strives for both simplicity and generality.
- SEM matrices are used for representing model parameters.
- Supscripts are used to capture {child, parent} linkages among variables within- and across- levels. For example, the \( \Lambda^{1,2} \) matrix involves dependent variables at level 1 and independent variables at level 2.
- Subscripts for parameters within matrices indicate {to, from} relationships between a pair of variables for the corresponding levels in the superscripts. For example \( \lambda_{m,n}^{u,v} \) indicates regression from \( n^{th} \) latent variable at level \( v \) to \( m^{th} \) observed dependent variable at level \( u \).
Linear mixed-effects model
The above model corresponds to a conventional LME
\[ Y=Xb + Zu + e, e \sim N(0,R),u \sim N(0,G). \]
LME model is xxM with a different name
\[ X_{ij} \equiv Z_{ij} \equiv \Lambda_{ij}, \] \[ b \equiv \alpha, \] \[ R_{ij} \equiv \Theta_{ij}, \text{&}\] \[ G_{ij} \equiv \Psi_{ij}. \]
xxM is a superset of LME model
- Most LME can be readily specified within xxM.
- xxM allows observed and latent variables at all levels.
- xxM offers more than one way of specifying the same model.
- xxM makes the specification of the \( R \) side of the LME very convenient.
- xxM allows non-block diagonal \( G \) matrices for some models.
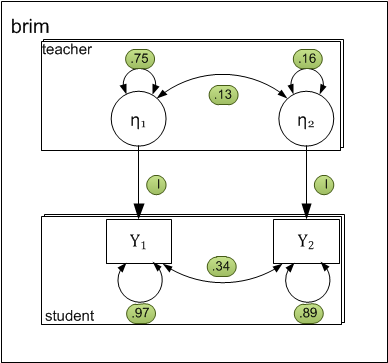
Path-diagram
The following two-level path diagram accurately represents all parameters. One- to-one correspondence between the diagram and the matrices makes it easy to specify the model.
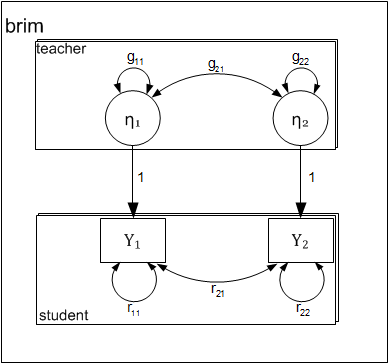
- Level-1 residual variances and covariance is represented by curved arrows labeled with the letter R.
- Level-2 variances and covariance among intercepts is represented by curved arrows labeled with the letter G.
- Each level-1 dependent variable \( (y_{pij}) \) is influenced by the corresponding level-2 intercept \( (\eta_{pj}) \). By definition the effect is fixed to 1.0.
Fitting bivariate random intercepts model in xxM
- Main Model
- One or more submodels
- Parameter matrices
Each of these objects is very simple. These model objects are constructed and added in a sequential fashion. Regardless of the complexity of the model, the process of constructing and adding model objects remains the same. An advantage of this approach is that complex models can be constructed by repeating the same steps multiple times. There are just three steps for constructing the model:
- Construct main model.
- Constuct submodels for each level and add to the main model.
- Construct parameter matrices and add to the main model or the appropriate sub-model.
Correspondingly, the actual model is constructed and estimated by invoking following commands:
xxmModel()xxmSubmodel()xxmWithinMatrix()xxmBetweenMatrix()
Matrices within a submodel are just a little differnt from across-level matrices. FInal command to estiamte the model is simplyxxmRun()
Library and Data
The first step is to load the `xxm` package and the `brim` dataset.
library(xxm)
data(brim.xxm, package = "xxm")Model
An n-level xxM model is composed of n submodels. The very first step in specifying an xxM model is to create a model object by invoking xxmModel(). The idea is to declare names of all levels.
brim <- xxmModel(levels = c("student", "teacher"))The function takes a single parameter aptly called levels and expects to receive a list of level names. Internally, xxM assigns level numbers \( (l = {1,2,\dotsc, L}) \) to each level declared in xxmModel(). The order of levels in the list is important. In this case, students are influenced by teachers. Hence, the student level must be declared before the teacher level.
The function creates an object called brim (i.e., bivariate random intercepts model). The left hand side is the name of the xxM model. The function actually returns a handle or a pointer to the object in memory. The choice of the name is arbitrary. However, it is better to use a short, but descriptive name, as this name will be used in all subsequent commands.
At this point, brim knows that there are two levels: student and teacher. Internally, the above invocation creates an object called brim with placeholders for the student and teacher submodels, as depicted below.

What is a level?
The term level is obvious in this simple case. Presumably students are nested within teachers. In multilevel modeling jargon, we have two levels with students hierarchically nested within teachers. The notion of a level in xxM is consistent with its conventional usage in the MLM literature.
A level represent any concrete or abstract set of entities across which some attribute is expected to vary. Very simply, a level involves multiple entities of some kind (e.g., students, situations, responses, occasions etc.) for whom there is an attribute or a variable (e.g., achievement) of interest. Each level may have its own set of observed and/or latent variables.
What are parent and child levels?
Levels are really quite nebulous in xxM. xxM uses a more general notion of parent-child relationship across levels to specify dependency structure. In this case, teacher is the parent level that influences student or the child level. More generally,
- Parent level includes observed or latent independent variables that influence the child level.
- Child level has observed or latent dependent variables influenced by the parent level.
The notion of parent-child relationships in xxM allows fairly complex models to be estimated without the complexities of managing subscripts for all levels simultaneously.
The definition of levels and in particular the idea of parent-child relationship implies two things:
- Directionality of influence: parent levels may influence child levels and not other way around.
- Order: Levels are ordered internally from low to high. A child level is at a lower level than a parent level. This order is implied in the levels argument of
xxmModel()
Submodels
Each level may have its very own complete SEM model with observed dependent and exogenous independent variables, latent variables, measurement model, and structural model involving all possible regressions (observed on observed, observed on latent, latent on observed, and latent on latent). Before we can begin to specify the actual model, we need to provide our model object- brim, with basic information about each level. This is accomplished by the xxmSubmodel() function.
brim <- xxmSubModel
brim <- xxmSubmodel(model = brim, level = "student", parents = c("teacher"), ys = c("y1",
"y2"), xs = , etas = , data = brim.student)The xxmSubmodel() function adds basic information about each level to our xxM model object, brim.
- model: The first parameter model, asks for the name of the
xxMobject to which this information is being added. - level: The second parameter identifies the level for the submodel. In this case, we are adding information about the student level.
- parents: The next parameter, parents, defines the nesting relationship involving students. Students are nested within teachers and the nesting is captured by the notion of parent and child levels in
xxM. In this case, the teacher level is a parent of the student level. If there were additional levels of nesting, these would be added to the list of parents as well. The following code provides an example with four levels:
parents = c("student", "teacher", "school", "neighborhood")- ys A list of observed dependent variables. In this case, we have two dependent variables for student (y1 and y2).
- xs A list of observed independent variables. There are no exogenous predictors at the student level.
- etas A list of latent dependent variables. There are no latent variables at the student level.
- data The final parameter, data, is for an R dataset with student data.
The corresponding submodel for the teacher level is:
brim <- xxmSubmodel(model = brim, level = "teacher", parents = , ys = , xs = , etas = c("eta1",
"eta2"), data = brim.teacher)The teacher level does not have a parent, nor does it have observed dependent or independent variables. The teacher level does have two latent variables. The latent variables represent random-intercepts of student level dependent variables (y1 and y2). If teachers were nested within a higher level such as school, the parents argument would be:
parents = c("school")How are datasets structured?
For two level data structures, a single dataset is adequate. However with complex dependent data-structures it is most convenient to provide data for each level separately. Each dataset must include information about how each observation at a lower level is linked to a unit at a higher level. In general, datasets may have three types of variables:
- One or more columns of IDs or variables with linking information. ID columns are mandatory.
- Zero or more columns of dependent variables corresponding to the list of ys. (Optional)
- Zero or more columns of independent variables corresponding to the list of xs. (Optional).
The student data has four columns student, teacher, y1 and y2. The first column is for the ID variable for the current level, in this case “student”. Student has a single parent: “teacher”. The ID columns must have the same name as the name of the corresponding level. Practically, it means that if in your dataset the ID variables are named SID and TID, these must be renamed to student and teacher.
The teacher dataset (teacher) has no observed variables, nor does it have any parents. Yet, it is necessary to have a dataset listing teacher IDs. The teacher data has a single column teacher.
A complete checklist to ensure that the data requirements are met:
- Each level must have a corresponding R dataset. To avoid confusion, the name of the dataset should match the level name.
- If a level does not have observed dependent or independent variables, the dataset would inclde a single column of level IDs.
- The first (1 + p) columns of a dataset include ID variables.
- First column is the ID column for the current level.
- The next p columns are the IDs for the parents of the current level.
- The names and order of the ID columns must match the corresponding level names declared in xxmModel.
- ID columns must be of type integer. R routinely converts categorical variables to a
factortype. - Dependent and independent variables must be of type numeric
- Use R command for examining structure of a dataset to ensure that the above requirements are met, e.g.
str(myLevel1Data)andstr(myLevel2Data).
So far we created a model object called brim by invoking xxmModel() and declared submodels for student and teacher by invoking xxmSubModel(). At this point, brim, is just a shell of the final model. The next logical step would be to specify the actual model, i.e., how observed and latent variables relate to each other. This is accomplished by defining parameter matrices.

Matrices
From an xxM perspective, the model is specified in terms of parameters and matrices associated with each level and links among variables across levels. This sounds complicated, but in reality we will simply repeat what we have already stated in previous sections:
What parameters are we estimating?
Parameters of interest in the above model specification are best specified as matrices. This allows complex models to be expressed succinctly. We begin by translating our scalar model formulation into xxM parameter matrices.
Within-student model matrices (Level 1)
At level-1, we only have variances and a covariance for the residuals. The residual covariance matrix is called the theta matrix (\( \Theta \)). The matrix is symmetric with three free parameters: two variances and a covariance \( (\theta_{12}= \theta_{21}) \).
\Theta^{1,1} =
\begin{bmatrix}
\theta_{1,1}^{1,1} & \\
\theta_{2,1}^{1,1} & \theta_{2,2}^{1,1}
\end{bmatrix}
\]
Within-teacher model matrices (Level-2)
At level-2, we have a covariance and variances among the latent variables, along with their means. Latent covariance and mean matrices are called \( \Psi \) (psi) and \( \alpha \) (alpha), respectively:
\Psi^{2,2}=
\begin{bmatrix}
\psi_{1,1}^{2,2} & \\
\psi_{2,1}^{2,2} & \psi_{2,2}^{2,2}
\end{bmatrix}
,
\alpha^{2}=
\begin{bmatrix}
\alpha_{1}^{2} \\
\alpha_{2}^{2}
\end{bmatrix}
\]
Across-level model matrices: From Teachers To Students ( Level 2 -> Level 1)
The teacher level latent variables influence student level observed variables. The coefficients matrix is \( \Lambda \) (lambda):
\Lambda =
\begin{bmatrix}
1 & 0\\
0 & 1
\end{bmatrix}
\]
In LISREL and xxM, \( \Lambda \) matrix is used to capture measurement relationship. In this case, level-2 latent variables are said to be measured by level-1 observed variables.
How are parameter matrices added to the model?
We have now defined four matrices that completely specify the underlying bivariate random-intercepts model. Once the model itself is clearly defined, the actual specification is trivial.There are just two commands for specifying parameter matrices:
brim <- xxmWithinMatrix(
)
brim <- xxmBetweenMatrix(
)Essentially, we want to add the above four matrices to complete the model.
xxmWithinMatrix()three times, once for the student level and then twice for the teacher level.xxmBetweenMatrix()will be called once connecting the teacher level to the student level. The following code fragment illustrates our intent.
brim <- xxmWithinMatrix(model = brim, level = "student", type = "theta")
brim <- xxmWithinMatrix(model = brim, level = "teacher", type = "psi")
brim <- xxmWithinMatrix(model = brim, level = "teacher", type = "alpha")
brim <- xxmBetweenMatrix(model = brim, parent = "teacher", child = "student", type = "lambda")Now we know the general procedure for adding a matrix to the model. Let us now examine how a parameter matrix to be added is actually constructed.
What are free and fixed parameters?
Note that the first three parameter matrices (\( \Theta \), \( \Psi \), and \( \alpha \)) are somewhat different from the last matrix (\( \Lambda \)). The first three matrices include model parameters that are to be freely estimated. In contrast, all four elements of the last matrix are fixed. We already know their values. This idea of free vs. fixed parameters is central in SEM. In essence, for each parameter we need to tell xxM if the parameter is to be estimated or if the parameter is to be fixed to some known value. For each parameter matrix, we need to define two separate matrices:
1. pattern matrix indicating the pattern of free (\( = 1 \)) or fixed (\( = 0 \)) parameters and
2. value matrix providing numeric values for fixed-parameters or start-values for free parameters.
It is easier done than said. We use a two part name including:
- Matrix type (\( \Theta \), \( \Psi \), \( \Lambda \) and \( \alpha \)).
- Matrix role (pattern or value ).
lambda_pattern <- matrix(c(0, 0, 0, 0), 2, 2)
lambda_value <- matrix(c(1, 0, 0, 1), 2, 2)\Lambda_{pattern} =
\begin{bmatrix}
0 & 0 \\
0 & 0
\end{bmatrix}
\] \[
\Lambda_{value} =
\begin{bmatrix}
1.0 & 0.0 \\
0.0 & 1.0
\end{bmatrix}
\]
All elements of pattern matrix for \( \Lambda \) are zero indicating that none of the parameters are free to be estimated. Instead, all parameters are to be fixed to some known values. The value matrix provides the corresponding values. The diagonal elements are to be fixed to 1.0, whereas the off-diagonal elements are to be fixed to 0.0. Compare the specification of \( \Lambda \) with that of the \( \Theta \) matrix:
theta_pattern <- matrix(c(1, 1, 1, 1), 2, 2)
theta_value <- matrix(c(1.1, 0.2, 0.2, 2.3), 2, 2)\Theta_{pattern} =
\begin{bmatrix}
1 & 1 \\
1 & 1
\end{bmatrix}
\] \[
\Theta_{value} =
\begin{bmatrix}
1.1 & 0.2 \\
0.2 & 2.3
\end{bmatrix}
\]
All four elements of the \( \Theta \) matrix are to be freely estimated. Hence all four elements in the pattern matrix are 1s. The value matrix provides start values. At this point, you may complain that you do not have any idea as to what these values may be. In general, almost any reasonable set of values will work. More specifically, for a residual covariance matrix, the following rules work very well in practice:
- Start values for the residual variances or the diagonal elements may be close to the observed variances of the respective variables.
- Start values for the residual covariances or the off-diagonal elements may be close to zero. Again, actual values do not matter much.
So far, we have described
- The four model matrices.
- Structure and meaning of pattern and value matrices.
Now we will see how these matrices are constructed in R and added to our xxM model object brim.
How do I construct and add matrices to the model?
We create pattern and value matrices for each of the four model matrices, and add these matrices to our model as described earlier:
Within-student model matrices
# Theta create
theta_pattern <- matrix(c(1, 1, 1, 1), 2, 2)
theta_value <- matrix(c(1.1, 0.2, 0.2, 2.3), 2, 2)
# Theta add
brim <- xxmWithinMatrix(model = brim, level = "student", type = "theta", pattern = theta_pattern,
value = theta_value)Within-teacher model matrices
# Psi create
psi_pattern <- matrix(c(1, 1, 1, 1), 2, 2)
psi_value <- matrix(c(0.1, 0.05, 0.05, 0.2), 2, 2)
# psi add
brim <- xxmWithinMatrix(model = brim, level = "teacher", type = "psi", pattern = psi_pattern,
value = psi_value)
# Alpha create
alpha_pattern <- matrix(c(1, 1), 2, 1)
alpha_value <- matrix(c(1.1, 2.1), 2, 1)
# Alpha add
brim <- xxmWithinMatrix(model = brim, level = "teacher", type = "alpha", pattern = alpha_pattern,
value = alpha_value)Teacher -> Student: Across level matrix
# Lambda create
lambda_pattern <- matrix(c(0, 0, 0, 0), 2, 2)
lambda_value <- matrix(c(1, 0, 0, 1), 2, 2)
# Lambda add
brim <- xxmBetweenMatrix(model = brim, parent = "teacher", child = "student", type = "lambda",
pattern = lambda_pattern, value = lambda_value)Run
If all went well above and xxM did not produce any error messages, then our model object brim has all the information it needs to estimate the model parameters. We can begin estimation by issuing a simple command:
brim <- xxmRun(brim)Code listing
xxM
This section presents annotated output of running the “brim.xxm.R” script at the R prompt. You can find the script in <r_library_directory\xxm\models\brim> directory. Running the script should reproduce the results. You may experiment by providing unreasonable start values.
Load xxM
xxM library needs to be loaded first. Data for the model must be available in the workspace.
library(xxm)
data(brim.xxm, package = "xxm")Construct R-matrices
For each parameter matrix, construct three related matrices:
- pattern matrix: A matrix indicating free or fixed parameters.
- value matrix: with start or fixed values for corresponding parameters.
- label matrix: with user friendly label for each parameter. label matrix is optional.
theta_pattern <- matrix(c(1, 1, 1, 1), nrow = 2, ncol = 2)
theta_value <- matrix(c(1, 0, 0, 1), nrow = 2, ncol = 2)
alpha_pattern <- matrix(1, 2, 1)
alpha_value <- matrix(c(0, 0), 2, 1)
psi_pattern <- matrix(c(1, 1, 1, 1), 2, 2)
psi_value <- matrix(c(0.1, 0, 0, 0.1), 2, 2)
lambda_pattern <- matrix(c(0, 0, 0, 0), 2, 2)
lambda_value <- matrix(c(1, 0, 0, 1), 2, 2)Construct model
xxmModel() is used to declare level names. The function returns a model object that is passed as a parameter to subsequent statements. Variable name for the return value can be anything. A
brim <- xxmModel(levels = c("student", "teacher"))## A new model was created.Add submodels
For each declared level xxmSubmodel() is invoked to add corresponding submodel to the model object. The function adds three types of information to the model object:
- parents declares a list of all parents of the current level.
- Level with the independent variable is the parent level.
- Level with the dependent variable is the child level.
- variables declares names of observed dependent (ys), observed independent (xs) and latent variables (etas) for the level.
- data R data object for the current level.
### Submodel: Student
brim <- xxmSubmodel(model = brim, level = "student", parents = c("teacher"), ys = c("y1",
"y2"), xs = , etas = , data = brim.student)## Submodel for level `student` was created.### Submodel: Teacher
brim <- xxmSubmodel(model = brim, level = "teacher", parents = , ys = , xs = , etas = c("eta1",
"eta2"), data = brim.teacher)## Submodel for level `teacher` was created.Add within-level matrices
For each declared level xxmWithinMatrix() is used to add within-level parameter matrices. For each parameter matrix, the function adds the three matrices constructed earlier:
- pattern
- value
- label (optional)
### Within-Matrix: Level :: Student, Type :: observed residual-covariance
brim <- xxmWithinMatrix(model = brim, level = "student", type = "theta", pattern = theta_pattern,
value = theta_value)
##
## 'theta' matrix does not exist and will be added.
## Added `theta` matrix.
### Within-Matrix: Level :: Teacher, Type :: latent residual covariance
brim <- xxmWithinMatrix(model = brim, level = "teacher", type = "psi", pattern = psi_pattern,
value = psi_value)
##
## 'psi' matrix does not exist and will be added.
## Added `psi` matrix.
### Within-Matrix: Level :: Teacher, Type :: latent means
brim <- xxmWithinMatrix(model = brim, level = "teacher", type = "alpha", pattern = alpha_pattern,
value = alpha_value)
##
## 'alpha' matrix does not exist and will be added.
## Added `alpha` matrix.Add across-level matrices
Pairs of levels that share parent-child relationship have regression relationships. xxmBetweenMatrix() is used to add corresponding regression matrices connecting the two levels.
- Level with the independent variable is the parent level.
- Level with the dependent variable is the child level.
For each parameter matrix, the function adds the three matrices constructed earlier:
- pattern
- value
- label (optional)
### Between-Matrix: Parent :: Teacher, Child :: Student, Type ::
### factor-loadings
brim <- xxmBetweenMatrix(model = brim, parent = "teacher", child = "student", type = "lambda",
pattern = lambda_pattern, value = lambda_value)##
## 'lambda' matrix does not exist and will be added.
## Added `lambda` matrix.Estimate model parameters
Estimation process is initiated by xxmRun(). If all goes well, a q&d summary of the results is printed.
brim <- xxmRun(brim)## ------------------------------------------------------------------------------
## Estimating model parameters
## ------------------------------------------------------------------------------
## 456.5781921615
## 454.3359125436
## 452.6051730894
## 451.0652787053
## 450.0531684945
## 448.6173091428
## 447.5320743183
## 447.1954369085
## 445.8740258784
## 445.7721481833
## 445.4488373694
## 445.4357270893
## 445.4338550574
## 445.4331653124
## 445.4331434919
## 445.4331407646
## 445.4331407251
## Model converged normally
## nParms: 8
## ------------------------------------------------------------------------------
## *
## 1: student_theta_1_1 :: 0.969 [ 0.000, 0.000]
##
## 2: student_theta_1_2 :: 0.338 [ 0.000, 0.000]
##
## 3: student_theta_2_2 :: 0.893 [ 0.000, 0.000]
##
## 4: teacher_psi_1_1 :: 0.752 [ 0.000, 0.000]
##
## 5: teacher_psi_1_2 :: 0.128 [ 0.000, 0.000]
##
## 6: teacher_psi_2_2 :: 0.159 [ 0.000, 0.000]
##
## 7: teacher_alpha_1_1 :: -0.107 [ 0.000, 0.000]
##
## 8: teacher_alpha_2_1 :: 0.278 [ 0.000, 0.000]
##
## ------------------------------------------------------------------------------
Estimate profile-likelihood confidence intervals
Once parameters are estimated, confidence intervals are estimated by invoking xxmCI(). Depending on the the number of observations and the complexity of the model, xxmCI() may take a long time to compute. xxmCI() also prints a summary of parameter estimates and CIS.
View results
A summary of results may be retrived as an R list by a call to xxmSummary(). The returned list has two elements:
- fit is a list with five elements:
- deviance is \( -2 Log Likelihood \) for the maximum likelihood fit function.
- nParameters is the total number of unique parameters.
- nObservations is the total number of observations across all levels.
- aic is Akaike’s Information Criterion or AIC computed as \( -2ll + 2*p \).
- bic is Bayesian Information Criterion or BIC computed as \( -2ll + p*\log(n) \).
- estimates is a single table of free parameter estimates
All xxM parameters have superscripts {child, parent} and subscripts {to, from}. xxM adds a descriptive parameter label if one is not already provided by the user.
xxmSummary(xm)## $fit
## $fit$deviance
## [1] 445.4
##
## $fit$nParameters
## [1] 8
##
## $fit$nObservations
## [1] 150
##
## $fit$aic
## [1] 461.4
##
## $fit$bic
## [1] 485.5
##
##
## $estimates
## child parent to from label estimate low high
## 1 student student y1 y1 student_theta_1_1 0.9691 0.670784 1.4741
## 2 student student y1 y2 student_theta_1_2 0.3381 0.090586 0.6823
## 3 student student y2 y2 student_theta_2_2 0.8935 0.618407 1.3589
## 8 teacher teacher eta1 eta1 teacher_psi_1_1 0.7524 0.293264 1.6633
## 9 teacher teacher eta1 eta2 teacher_psi_1_2 0.1283 -0.156114 0.5437
## 10 teacher teacher eta2 eta2 teacher_psi_2_2 0.1592 0.001000 0.5548
## 11 teacher teacher eta1 One teacher_alpha_1_1 -0.1073 -0.529914 0.3154
## 12 teacher teacher eta2 One teacher_alpha_2_1 0.2781 0.002623 0.5537
Free model object
xxM model object may hog a significant amount of RAM outside of R’s workspace. This memory will automatically be released, when the workspace is cleared by a call to rm(list=ls()) or at the end of the R session. Alternatively, it is recommended that xxmFree() may be called to release the memory.
brim <- xxmFree(brim)Proc Mixed
Proc Mixed data = brim covtest;
CLASS student teacher vars;
MODEL y = vars / solution noint;
RANDOM vars / subject = teacher type = un;
REPEATED vars/ subject = student(teacher) type = un;
RUN;